Seventh post in a series of posts written by members of the Qurit Team. Written by Asim Shrestha.
Introducing RT-UTILS

DICOM and RT Structures are the primary means in which medical images are stored. Because of this, it is important to be able to translate data in and out of RT Structs. Although there are well documented mechanisms for loading RT Structures within Python, we found that creating such files ourselves proved difficult. Existing tools felt complicated and clunky and so we set out to make our own library. Introducing RT-Utils, a lightweight and minimal python package for RT Struct manipulation.
What kind of manipulation is possible?
When we say manipulation, we mean that users are able to both load in existing RT Struct files or create new RT Structs given a DICOM series. After doing so, they can perform the following operations:
- Create a new ROI within the RT Struct from a binary mask. Optionally, one can also specify the color, name, and description of the RT Struct.
- List the names of all the ROIs within the RT Struct.
- Load ROI mask from the RT Struct by name.
- Save the RT Struct by name or path.
How are DICOM headers dealt with?
Our approach with headers are simple. We first add all of the headers required to make the RT Struct file a valid RT Struct. After this, we transfer all of the headers we can from the input DICOM series such as patient name, patient age, etc. Finally, as users input ROIs within the RT Struct, they will be dynamically added to the correct sequences within the RT Struct.
Installation
pip install rt-utils
After installation, you then import RTStructBuilder and either create a new RT Struct or load an existing RT Struct. Below are examples of both of these. Pay attention to the comments in the code snippets to get a better idea of what is going on.
Adding a ROI mask
The ROI mask format required for the add_roi method is a 3D array where each 2D slice of the array is a binary mask. Values of one within the mask indicate that the pixel is a part of the ROI, for a given slice. There must be as many slices within the array as there are slices within your DICOM series. The first slice in the array corresponds to the smallest slice location in the DICOM series, the second slice corresponds to the second slice location, etc.
Creating new RT Structs
from rt_utils import RTStructBuilder
rtstruct = RTStructBuilder.create_new(dicom_series_path="./testlocation")
# ...
# Create mask through means such as ML
# ...
# Add the 3D mask as an ROI.
# The colour, description, and name will be auto generated
rtstruct.add_roi(mask=MASK_FROM_ML_MODEL)
# Add another ROI, this time setting the color, description, and name
rtstruct.add_roi(
mask=MASK_FROM_ML_MODEL,
color=[255, 0, 255],
name="RT-Utils ROI!"
)
rtstruct.save('new-rt-struct')
view rawrt-utils-new.py hosted with ❤ by GitHub

Loading and viewing existing RT Structs
from rt_utils import RTStructBuilder
import matplotlib.pyplot as plt
# Load existing RT Struct. Requires the series path and existing RT Struct path
rtstruct = RTStructBuilder.create_from(
dicom_series_path="./testlocation",
rt_struct_path="./testlocation/rt-struct.dcm"
)
# View all of the ROI names from within the image
print(rtstruct.get_roi_names())
# Loading the 3D Mask from within the RT Struct
mask_3d = rtstruct.get_roi_mask_by_name("ROI NAME")
# Display one slice of the region
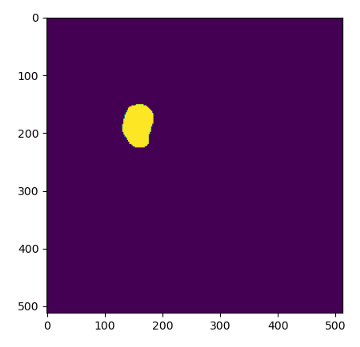
first_mask_slice = mask_3d[:, :, 0]
plt.imshow(first_mask_slice)
plt.show()
view rawrt-utils-loading.py hosted with ❤ by GitHub
Bonus: Dealing with holes in masks and ROI’s
Often times, DICOM viewing tools such as MIM will automatically detect inner and outer contours and draw holes accordingly. This is not always the case however and sometimes you will be required to create pin holes within your ROI so that your ROIs are displayed properly. This process is described in this link.

If this is the situation you are under, we have another optional parameter that can deal with this for you. When you are adding ROIs to your RT Struct, simply set use_pin_hole=True like as the example below.
rtstruct.add_roi(mask, use_pin_hole=True)
view rawrt-utils-pinhole.py hosted with ❤ by GitHub
