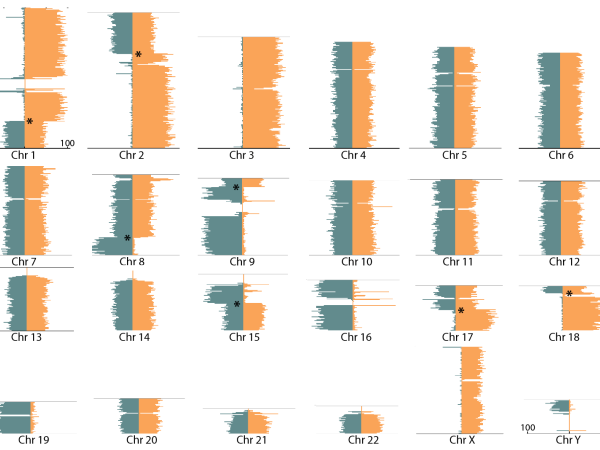
The SCORE will typically produce at least 20 good quality single-cell Strand-seq libraries with 10 million read pairs for each sample. Higher numbers of good libraries and greater sequencing depth are often obtained or a higher thresholds can be made upon request.
The above sequencing is sufficient for Strand-seq applications such as chromosome-length phasing of haplotypes for POAga and large structural variation detection, but genome assembly studies may require deeper genome coverage. Sample and sequencing costs may vary depending on the desired number of libraries, their desired sequencing depth and the possibility to make and sequence libraries concurrently with samples from different experiments.
The standard Strand-seq package includes cell culture with BrdU, preparation and fluorescence-activated cell sorting (FACS) of fixed nuclei, library preparation using the cellenONE, size selection and quality control of libraries, AVITI Next-generation DNA sequencing, and Strand-seq data processing. For other single-cell whole genome sequencing (WGS) applications, cell culture, nuclei preparation, and FACS can be omitted to reduce costs. Whole blood (2 x 6 mL sodium heparin) or cultured cell lines (frozen in cryovials or in culture media) are acceptable for submission. Alternatively, Coriell catalogued cell lines can be order by the SCORE. The SCORE will provide FastQ files and basic sequencing metrics upon request. We can offer assistance using Strand-seq bioinformatics tools.
Strand-seq Data Production Package
· Cell Culture, Nuclei Preparation & FACS - $200 / sample
· Nanolitre Library Preparation & QC - $100 / sample (in a batch of ~15-20 samples)
· AVITI Sequencing and Data Processing - $1770 (2 x 75 bp) or $2760 (2 x 150 bp) / run (multiplexed pool of ~15-20 samples)
· Labour - $40 / hour
· A surcharge is applied for external users (25%) and corporate users (40%) of the total cost, and may include an additional $500 administrative fee per invoice billed quarterly.
Typical cost per sample (20+ good cells, > 10 million read pairs): $500-1000 depending on pooling of libraries and sequencing strategy.