My laboratory explores regulatory mechanisms in organ development, in particular the liver and heart valves. We are using state of the art genomic technologies to shed new light on developmental gene networks by using whole tissue and single cell analyses of gene expression, transcription factor function and epigenetic mechanisms.
HOW DO WE MAKE A LIVER?
The liver is a fascinating organ which controls a wide-array of homeostatic processes in the body including detoxification of metabolites and chemicals, glucose metabolism, synthesis of lipids, production of serum proteins, and bile production. Many people will experience liver disease in their lifetime but, unlike most tissues, this organ retains the ability to regenerate itself when damaged. Despite this ability, chronic liver disease is on the rise. Obesity is the leading cause of liver disease, with over-exposure to common chemicals or medications and viral infections such as hepatitis, being major contributors. Regardless of the initiating factor, chronic liver disease can cause permanent liver damage and scarring that will progress to cirrhosis, poor liver function and can advance to cancer. Currently, there is no cure for cirrhosis. When it is no longer possible to manage the disease, a liver transplant is required, but demand far exceeds available donor tissue. A full understanding of how a liver develops in the embryo will contribute to better ways of treating disease, as well as the generation of alternative sources for liver transplantation.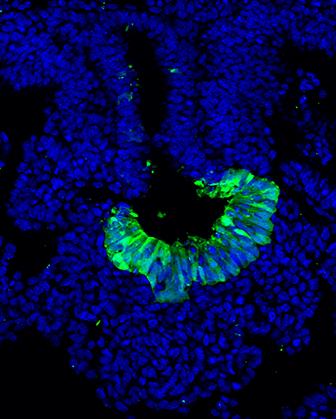
Liver formation initiates as an epithelial bud that evaginates from the foregut endoderm in response to signals from neighbouring mesoderm. This bud grows and, in association with mesenchyme, endothelium and blood cells, differentiates to form the functional organ. We seek to understand the regulatory processes in early liver formation, including spatial signalling and cellular differentiation, in both mouse and human pluripotent stem cells.
We have two main questions that we are currently exploring:
What are the mechanisms through which transcription factors and epigenetic modifications cooperate to regulate tissue development?
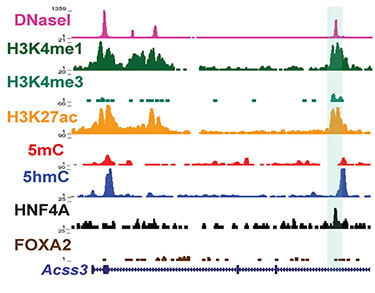 We are using the mouse and human pluripotent stem cells to study the transcriptional networks that control differentiation. Our studies centre on two transcription factors essential for hepatocyte development and function, Hepatocyte Nuclear Factor 4A and FoxA2. Our work shows that these factors have distinct and overlapping functions in development and in the adult. However, it is unclear how the embryonic and adult specific functions are controlled. We are studying the associated epigenetic mechanisms to better understand how these transcriptions factors function to drive differentiation. By studying how sequence-specific, DNA-binding factors regulate transcriptional networks, interact with histone and DNA modifications, and with the enzymes that catalyze these modifications, we can explore how differentiation steps are controlled in specific ways.
We are using the mouse and human pluripotent stem cells to study the transcriptional networks that control differentiation. Our studies centre on two transcription factors essential for hepatocyte development and function, Hepatocyte Nuclear Factor 4A and FoxA2. Our work shows that these factors have distinct and overlapping functions in development and in the adult. However, it is unclear how the embryonic and adult specific functions are controlled. We are studying the associated epigenetic mechanisms to better understand how these transcriptions factors function to drive differentiation. By studying how sequence-specific, DNA-binding factors regulate transcriptional networks, interact with histone and DNA modifications, and with the enzymes that catalyze these modifications, we can explore how differentiation steps are controlled in specific ways.
How do cell-cell communications drive regulatory networks required for normal liver formation?
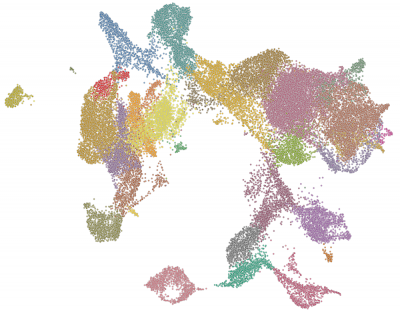
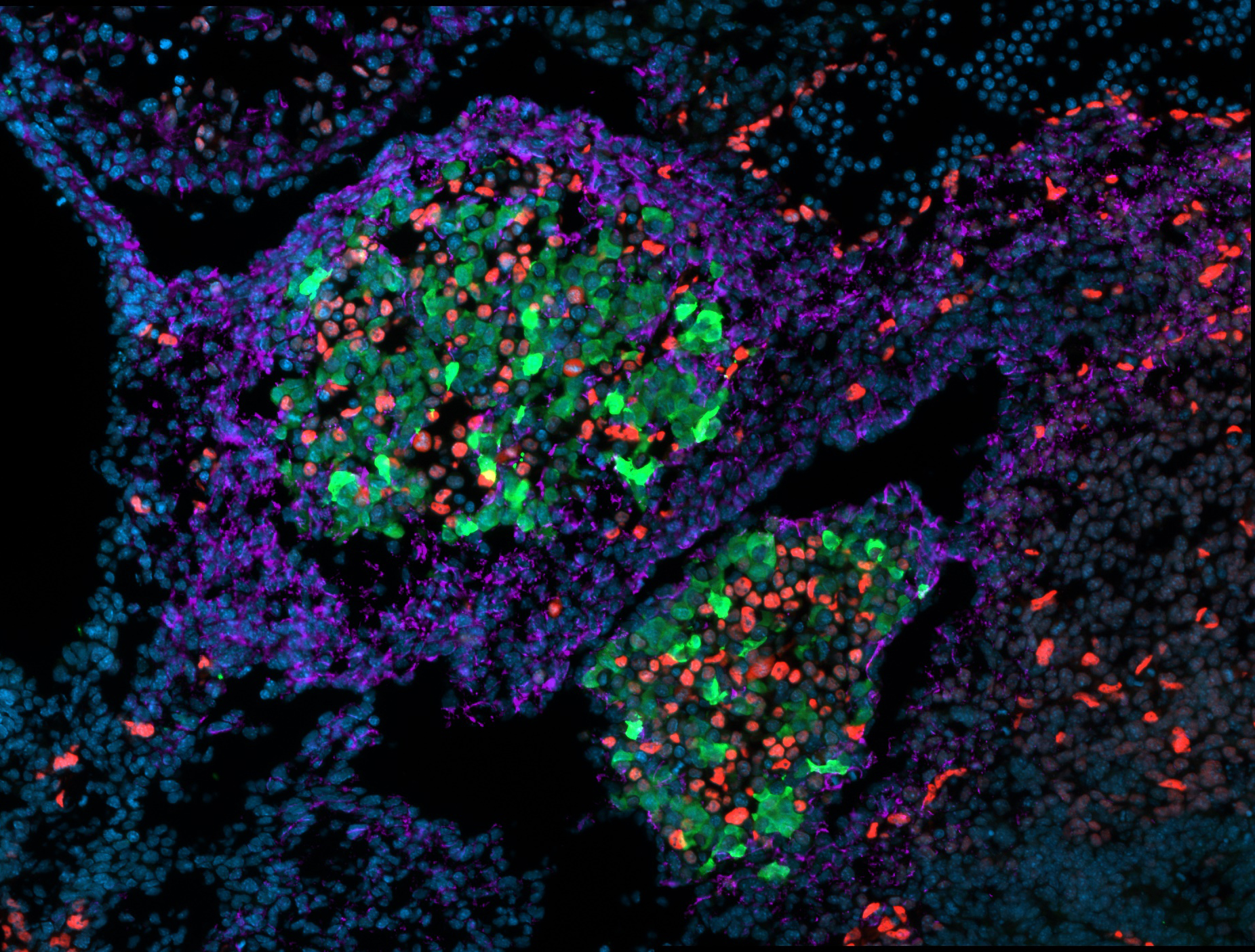 Although the primary functional cells in the liver, hepatocytes and cholangiocytes (bile duct), arise from the endoderm, other types of cells contribute to the development of a normal liver through cell-cell communication including endothelial, mesenchyme and blood cells. Cell-cell communications are important for proper liver development. However, we know very little about their role in hepatocyte and cholangiocyte differentiation or in liver morphology. We are using single cell and genome-wide analysis to explore the diversity of cells in the embryonic liver.
Although the primary functional cells in the liver, hepatocytes and cholangiocytes (bile duct), arise from the endoderm, other types of cells contribute to the development of a normal liver through cell-cell communication including endothelial, mesenchyme and blood cells. Cell-cell communications are important for proper liver development. However, we know very little about their role in hepatocyte and cholangiocyte differentiation or in liver morphology. We are using single cell and genome-wide analysis to explore the diversity of cells in the embryonic liver.
HOW DO WE MAKE HEART VALVES?
 Congenital heart malformations are the most common form of birth defects, with many of these involving the heart valves. In the embryonic heart, the valves develop in the region between the atria and the ventricles, known as the atrioventricular canal (AVC) and in the region where blood flows into the major arteries, known as the outflow tract (OFT). Valve formation initiates in these regions when endothelial cells which line of the interior of the heart, undergo endothelial-to-mesenchymal transition (EMT) to form endocardial cushions. The cushions undergo remodeling through cellular differentiation to form the valve leaflets and the valve interstitial cells secrete the extracellular matrix that provides the mechanical strength and flexibility to the valve leaflets. Very little is known about valve remodelling during development and how these processes are altered in valve disease. Understanding development may lead to alternative interventions in valve disease.
Congenital heart malformations are the most common form of birth defects, with many of these involving the heart valves. In the embryonic heart, the valves develop in the region between the atria and the ventricles, known as the atrioventricular canal (AVC) and in the region where blood flows into the major arteries, known as the outflow tract (OFT). Valve formation initiates in these regions when endothelial cells which line of the interior of the heart, undergo endothelial-to-mesenchymal transition (EMT) to form endocardial cushions. The cushions undergo remodeling through cellular differentiation to form the valve leaflets and the valve interstitial cells secrete the extracellular matrix that provides the mechanical strength and flexibility to the valve leaflets. Very little is known about valve remodelling during development and how these processes are altered in valve disease. Understanding development may lead to alternative interventions in valve disease.
How do transcriptional networks drive heart valve formation?
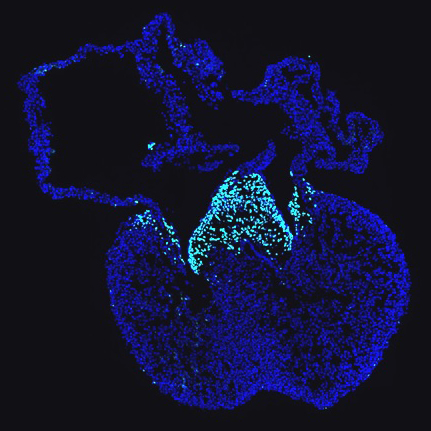 The transcription factor, SOX9, is essential to several aspects of valve development, but it also important in the development of many other tissues. Our work has shown that SOX9 plays a central role in regulating the transcriptional networks that drive valve differentiation. Moreover, SOX9 may function as a pioneer factor to establish gene expression necessary for normal development. We are using SOX9 as a window to understand the transcriptional and epigenetic changes that occur during valve development and how SOX9 functions to regulate distinct embryonic differentiation programs.
The transcription factor, SOX9, is essential to several aspects of valve development, but it also important in the development of many other tissues. Our work has shown that SOX9 plays a central role in regulating the transcriptional networks that drive valve differentiation. Moreover, SOX9 may function as a pioneer factor to establish gene expression necessary for normal development. We are using SOX9 as a window to understand the transcriptional and epigenetic changes that occur during valve development and how SOX9 functions to regulate distinct embryonic differentiation programs.